Polyphosphates, Polyphosphatase Activity and Stress Tolerance of Knockout Mutants Ppn1 and Ppn2 of Saccharomyces cerevisiae
- Affiliations
-
- 1. G.K. Skryabin Institute of Biochemistry and Physiology of Microorganisms, the Federal Research Center "Pushchino Scientific Center for Biological Research of the Russian Academy of Science
- Published:
- 2024-11-07
- Keywords:
- Polyphosphatase, polyphosphate, stress, PPN1, PPN2, mitochondria, Saccharomyces cerevisiae
Abstract
In this work, using the commercially available Saccharomyces cerevisiae knockout mutants (parent strain BY4741) in the PPN1 and PPN2 genes encoding polyphosphatases, the authors have shown that each of the two single mutations leads to the acquirement of similar physiological effects at the stationary stage of growth on glucose, namely the increased resistance to manganese and peroxide stresses and a higher level of long-chained polyphosphates. The increase in long-chain polyphosphate levels is more pronounced in the case of a knockout mutation in the PPN2 gene. The authors suggest, that the increased stress resistance of ∆ppn1 and ∆ppn2 strains is associated with the increase in the level of long-chained polyphosphate. The ∆ppn1 mutant cells did not differ in growth parameters in media with ethanol or glucose or the in the content of polyphosphates in mitochondria compared to the parent strain. The ∆ppn2 mutant had a longer lag phase in the transition to ethanol consumption. The data obtained support the idea that the polyphosphatases Ppn1 and Ppn2 are responsible for the regulation of the polyphosphate chain length, and polyphosphates and their metabolism are important for stress adaptation in yeast.
Full text
Introduction
Inorganic polyphosphate (polyP), a linear polymer of orthophosphoric acid, is present in all living cells from archaea to mammals. At present, an understanding has emerged about the participation of polyP and polyP metabolism enzymes in various processes regulating the vital activity of eukaryotes. [1-3]. The yeast can accumulate high amounts of polyP [4]. The knowledge on the role of these polymers in yeast extends from phosphate and energy reserve [4] to the key role in cell cycle regulation [5] and enzyme activity modification [6-8]. PolyP may influence cell physiology through non-enzymatic polyphosphorylation of lysine in proteins including DNA topoisomerase I, Nsr1, and ribosome biogenesis factors [7]. Yeast PolyP is involved in gene expression repression, presumably through the subtelomeric inhibition pathway of TORC2 [9].
One approach to understanding the pathways of polyP involvement in various cellular processes is to study the physiological characteristics of knockout mutations in genes encoding enzymes that metabolize polyP. Four polyphosphatases have been identified and characterized in Saccharomyces cerevisiae [10-14]. Among them, Ppn1 and Ppn2 are primarily responsible for the polyP level and chain length [10-13]. Both Ppn1 and Ppn2 display an endopolyphosphatase activity (polyphosphate depolymerase, EC 3.6.1.10.) and cleave long polyP chains into shorter ones:
PolyPn+H2O→polyPm (m<n)
Ppn1 also displays an exopolyphosphatase activity (polyphosphate phosphohydrolase, EC 3.6.1.11) and cleaves orthophosphate from the polyP chain end:
PolyPn+H2O→PolyPn-1+Pi
In addition, Ppn1 cleaves the terminal phosphate from nucleoside 5’ tetraphosphate and desoxynucleoside triphosphate [14]. Both Ppn1 and Ppn2 are able to split polyP from polyP-lysine phosphorylated proteins [7]. These activities expand the possibilities for involving these polyphosphates in regulatory processes. The Ppn1 and Ppn2 enzymes differ in cellular localization: Ppn2 is localized in the vacuolar membrane [13]; Ppn1 is also localized mainly in vacuoles but is observed in the cytoplasm under phosphate surplus [15] and is supposed to be responsible for exopolyphosphatase activities in the mitochondrial membrane and the nucleus [16].
The effects of knockout mutations in PPN1 and PPN2 genes on polyP metabolism varied depending on the growth stage and culture conditions. The ∆ppn1 mutant grown in minimal medium containing 2% glucose, 0.5% (NH4)2SO4, 7.35 mM phosphate, trace elements, salts and vitamins showed an increase in polyP levels and chain length, as well as growth inhibition and decreased cell viability at the stationary growth stage [11]. The polyP level increased in the mitochondria and vacuoles of the Δppn1 mutant and the polyP chain length increased in mitochondria, vacuoles and cytoplasm when the yeast cells were cultivated in the YPD medium [16]. The polyP levels in the Δppn1, Δppn2 and Δppn1Δppn2 mutants did not vary compared to the parent strain in cells cultivated in the Sc minimal medium [13]. Both mutations led to an increase in polyP chain length [7, 13]. It was concluded that polyphosphatases Ppn1 and Ppn2 make a key contribution to the regulation of polyP content and chain length in the cells of S. cerevisiae [7, 13].The other effects of PPN1 knockout were also observed. The Δppn1 mutant strain had reduced viability at the stationary growth stage [11] and was unable to grow on lactate and ethanol [17]. The normal cell cycle progression was disturbed and the time of DNA replication increased 2-fold in the double mutant Δppn1Δppx1 [5]. Differences in the genetics constructions of the mutants and in the culture conditions make it difficult to compare the literature data on the effect of PPN1 and PPN2 knockout on polyP and other physiological properties of yeast cells.
Therefore, the present work was aimed at studying the effects of Δppn1 and Δppn2 knockout mutations using standard commercial S. cerevisiae mutants on the following physiological properties: (i) the growth on glucose-and ethanol-containing media, (ii) the content and chain length of different polyP fractions, (iii) the polyphosphatase activities in some subcellular fractions, including mitochondria, and (iiii) resistance to stress caused by peroxide and toxic concentrations of manganese and cadmium.
Materials and methods
The S. cerevisiae wild-type (WT) strain BY4741 (MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) and BY4741-derived mutant strains (∆ppn1 and ∆ppn2) were obtained from the Dharmacon collection (https://horizondiscovery.com/en/non-mammalian-research-tools/products/yeast-knockout). Yeast cultures were maintained on a YPD agar medium containing 2% peptone (Pronadisa, Spain), 1% yeast extract (Pronadisa, Spain), 2% glucose, and 2% agar. The inoculum was grown in a liquid YPD medium. Cells for the experiments were cultured in a YPD medium and aYPEt, medium containing 1% ethanol (YPEt) instead of glucose. The inoculum was introduced into the medium to an absorption of ~0.065. The strains were cultivated at 29°C on shakers in flasks with 200 ml of the medium and a at stirring speed of 145 rpm in case of YPD medium and in flasks with 50 ml and a stirring speed of 250 rpm in case of YPEt medium. To plot the growth curves of the strains, the aliquots of cultures were taken at certain time intervals, the absorption was measured at λ=600 nm in a 3.07-mm cuvette with a KFK-3 photometer (ZOMZ, Russia). The biomass was collected at the stationary growth stage (24 h for YPD and 60 h for YPEt) by 10 min centrifugation at 4 000 g, washed thrice with cold (4°C) distilled water, weighed, frozen at -20°C, and used for polyP analysis.
Five fractions of polyP (acid-soluble polyP1, salt-soluble polyP2, alkali-soluble polyP3 and polyP4, and acid-insoluble polyP5) were extracted according to [18]. Phosphate (Pi) was determined colorimetrically [19]. The amount of polyP was normalized to the wet weight of biomass samples, standardized by centrifugation at 5 000 g for 15 min.
The exopolyphosphatase and endopolyphosphatase activities were measured in cell-free extract and crude membrane fraction. Yeast cells were harvested and washed twice with distilled water at 5.000 g for 10 min; spheroplasts were obtained as described [14]. Spheroplasts were lysed in a glass Potter homogenizer in 0.1 M sorbitol, 25 mM Tris-HCl, pH 7.2, supplemented with 0.5 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich, USA) and centrifuged at 5.000 g for 10 min. The supernatant was collected and centrifuged at 15 000 g for 60 min. The supernatants were cell-free extracts. The pellets (crude membrane fraction) were re-suspended in 25 mM Tris-HCl, pH 7.2.
The exopolyphosphatase activity was determined at 30°C by phosphate (Pi) formation rate determined colorimetrically [14]. The incubation medium contained 50 mM Tris-HCl, pH 7.2, 2 mM MgSO4, 200 mM NH4Cl, and 2.5 mM polyP (as Pi) with the average chain lengths of 208 phosphate residues (polyP208) (Monsanto, USA). The amount of the enzyme forming 1 µmole Pi in 1 min was taken as a unit of activity (U). The endopolyphosphatase activity was analyzed by a decrease in polyP208 chain length. The incubation medium contained 50 mM Tris-HCl, pH 7.2, 200 mM NH4Cl, 2 mM MgSO4, 9 mM polyP208 (as Pi), and cell-free extract or crude membrane fraction samples. Incubation time was 30 min at 30°. The reaction was stopped by adding HClO4 to a concentration of 0.5 M. Further sample preparation and electrophoresis in 30% PAAG with 7 M urea were performed as described [14].
BSA was used as a standard for protein assay with Bradford reagent (Thermo Fisher, USA) according to the manufacturer’s instructions.
Mitochondria were isolated by differential centrifugation according to [20] with modifications. To obtain mitochondria from glucose-grown cells, the cells were cultivated in YPD medium for 24 h. To obtain mitochondria from the cells cultivated with ethanol, cells were pre-grown in the YPD medium (0.5% yeast extract, 1% peptone, 1% glucose) in flasks with 200 ml of the medium on a shaker at 145 rpm for 24 h, precipitated, washed with water and transferred to the YPEth medium (0.5% yeast extract, 1% peptone, 1% ethanol) in flasks with 50 ml of the medium in an amount of 0.5 g of crude biomass per flask. Cultivation was carried out for 10 h at 29°C and 250 rpm. Spheroplasts were obtained as described in [16]. In the case of glucose-grown cells, spheroplasts were lysed in glass Potter-Elvehjem homogenizer in a buffer containing 20 mM Tris-Hcl, pH 6.8, 1 mM PMSF, 1 mM EDTA, 4 mM MgSO4, 0.02% bovine serum albumin, 0.05% concanavalin A. Afterwards, an equal volume of the same buffer but with 1.2 M sorbitol was quickly added with stirring. The homogenate was centrifuged at 3000 g for 10 min. The resulting supernatant was centrifuged at 10 000 g for 10 min. The precipitate was homogenized with a Teflon pestle in 0.6 M sorbitol buffer for 1 min and centrifuged at 2500 g for 10 min. Mitochondria from the supernatant obtained after the second low-speed centrifugation were precipitated at 10 000 g for 10 min.
In case of ethanol-grown cells, spheroplasts were destroyed in a buffer already containing 0.6 M sorbitol and without the second step of homogenization.
The ATPase activity was determined by the rate of Pi release at 30 °C for 30 min in 1 mL of the reaction mixture containing 1 mM MgSO4 and 1 mM ATP in 50 mM Tris-HCl at pH 8.5 in the presence and absence of 5 mM NaN3. Pi was assayed as described earlier [14].
The exopolyphosphatase activity was determined by the rate of Pi release at 30 °C for 30 min in 1 ml of the reaction mixture containing 2.5 mM MgSO4 and 1 mM polyP188 (Monsanto, USA) in 50 mM Tris-HCl at pH 7.2.
For polyP extraction, mitochondrial preparations were treated at 0 °C with 1 M HClO4 for 15 min under stirring and centrifuged at 10 000 g for 10 min. Pi and polyP content after the treatment with 1 M HCl at 100 оC for 10 min were measured in the supernatant as described above.
To determine the sensitivity to peroxide and heavy metal ions, yeast samples normalized by cell concentration (0.5×107 cell/mL) were added into the wells of sterile plates containing the YPD medium and different concentrations of Cd(CH3COO)2×2H2O, MnSO4, or hydrogen peroxide, cultured for 24 h, and the culture absorbance was measured at 594 nm with an EFOS photometer (Sapphire, Russia). The initial cell concentration was determined by counting in a standard Goryaev chamber.
The experiments were repeated three times, and the results are presented as the mean with standard deviation (Excel). Statistical significance was assessed relative to the parental wt strain data with the help of Student's t-test using standard Excel software. The experiments with electrophoresis were repeated twice and the typical pictures are presented.
Results
3.1. Growth on glucose and ethanol
The parent strain (wt) and mutant strains were cultivated in media containing 3 mM Pi. No growth defects were observed during cultivation of the ∆ppn1 and ∆ppn2 strains in the YPD medium (Fig. 1). When cultured in YPEth medium with 1% ethanol, wt and ∆ppn1 strains were similar in growth, whereas ∆ppn2 had a longer lag phase .(Fig. 1).
3.2. PolyP content and chain length
We have compared the content of polyP of different fractions in the cells of wt and mutant strains grown on glucose and ethanol in the stationary stage cells (Fig. 2A). No statistically significant differences in polyP content in various fractions were found between wt and ∆ppn1 cells both when cultivated in glucose- or ethanol-containing media (Fig. 2A).
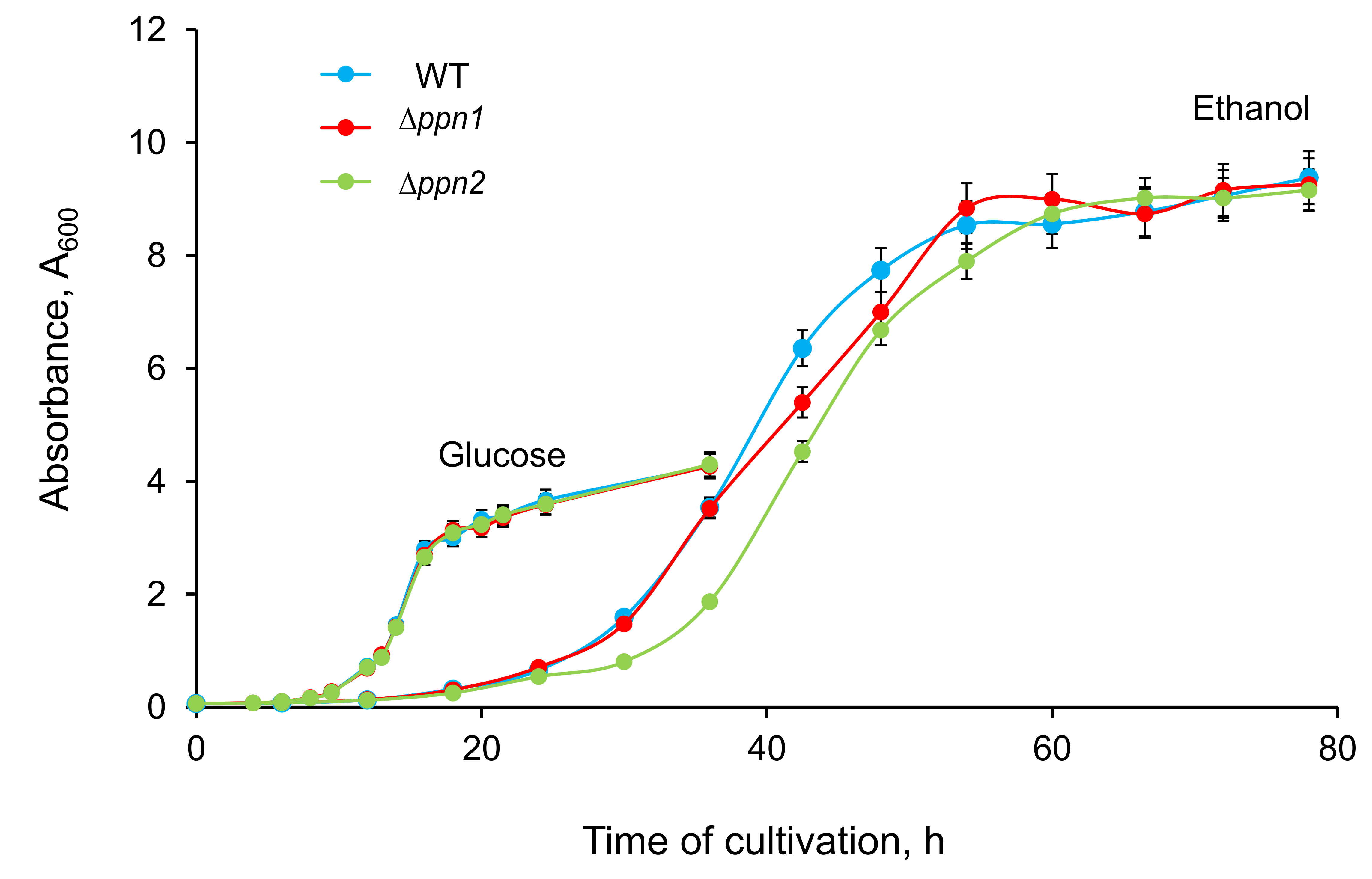
Figure 1. Growth curves of WT, ∆ppn1 and ∆ppn2 strains of S. cerevisiae in the YPD (glucose) and YPEth (ethanol) media
In the cells of the ∆ppn2 mutant, the content of relatively short-chained polyP1 and polyP2 decreased, while the content of longer-chained polyP3 and polyP5 increased independently of the carbon source used (Fig. 2A).
We also estimated the chain length of polyP of isolated fractions by electrophoresis using the polyP preparations obtained from glucose-grown cells (Fig. 2B).
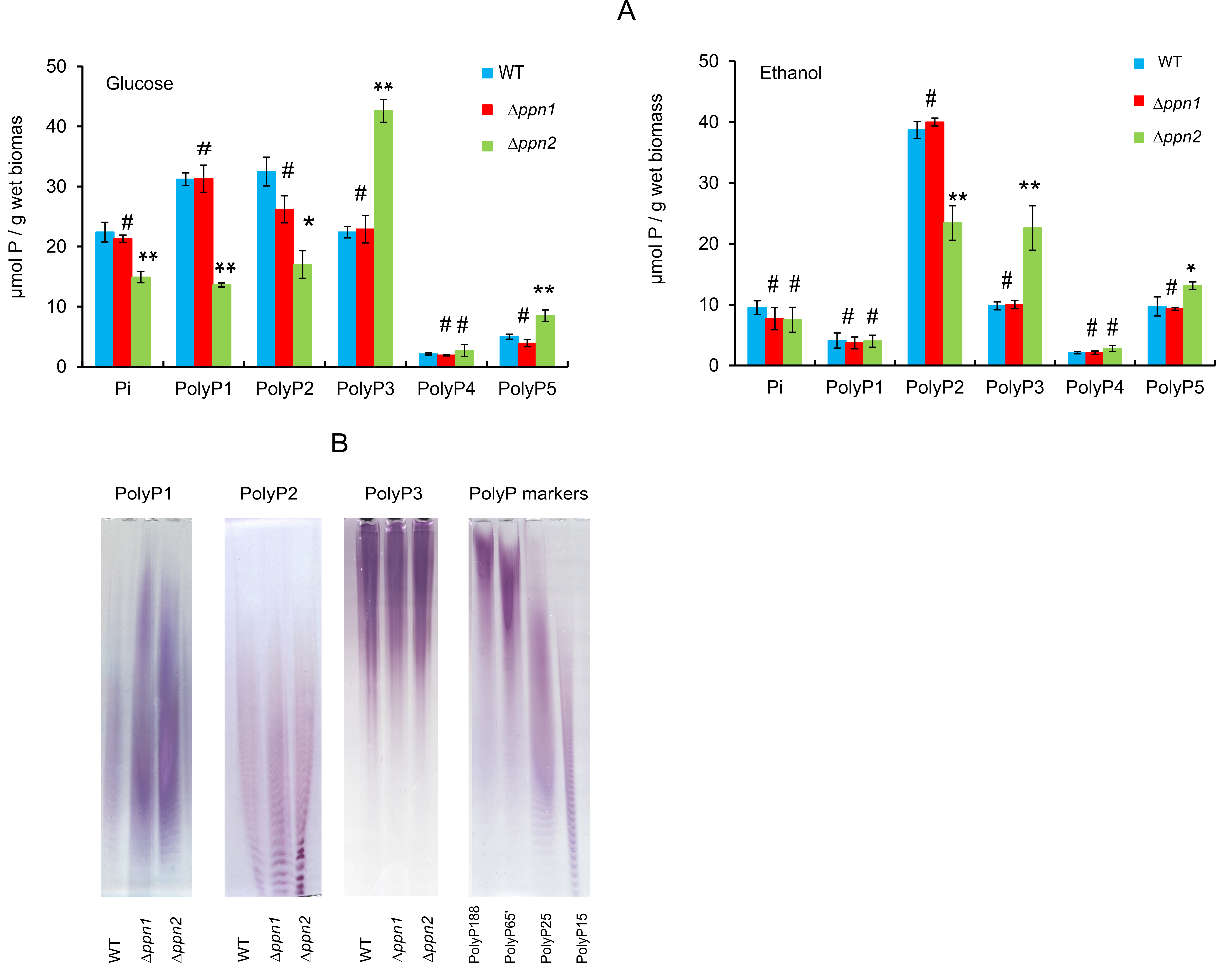
Figure 2. A - Pi and polyP content in the cells of S. cerevisiae strains cultivated in the YP media with glucose or ethanol up to the stationary growth stage. Statistical significance was assessed against wt with two tailed Student's t-test: * p < 0.05, ** p < 0.01, #-n.s. Statistical significance was assessed relative to the data for the parental wt strain with the help of Student's t-test using the standard Excel program: * p < 0.01, ** p < 0.001, # - the difference is statistically insignificant. B – the elecrophoregram of the preparations of polyP fractions polyP1, polyP2 and polyP3 obtained from the cells grown in the medium with glucose; polyP15-188 were preparations of commercial polyphosphates with average chain lengths from 15 to 188 phosphate residues (Sigma-Aldrich, USA)
The chain lengths of polyP2 and polyP3 were similar in the preparations obtained from all three strains. The chain length of polyP1 increased in case of both ∆ppn1 and ∆ppn2 strains. The increase in the proportion of long-chained polyP was most pronounced in the cells of the ∆ppn2 strain.
3.3. Polyphosphatase activities in subcellular fractions
We compared polyphosphatase activities in preparations of the soluble fraction (cell-free extract) and the crude membrane fraction for the three strains studied (Fig. 3).
As expected, in the crude membrane fraction, the ∆ppn1 mutant, there was a significant decrease in both exo- and endopolyphosphatase activities. In this fraction, the ∆ppn2 mutant also showed a decrease in endopolyphosphatase activity (Fig. 3B).
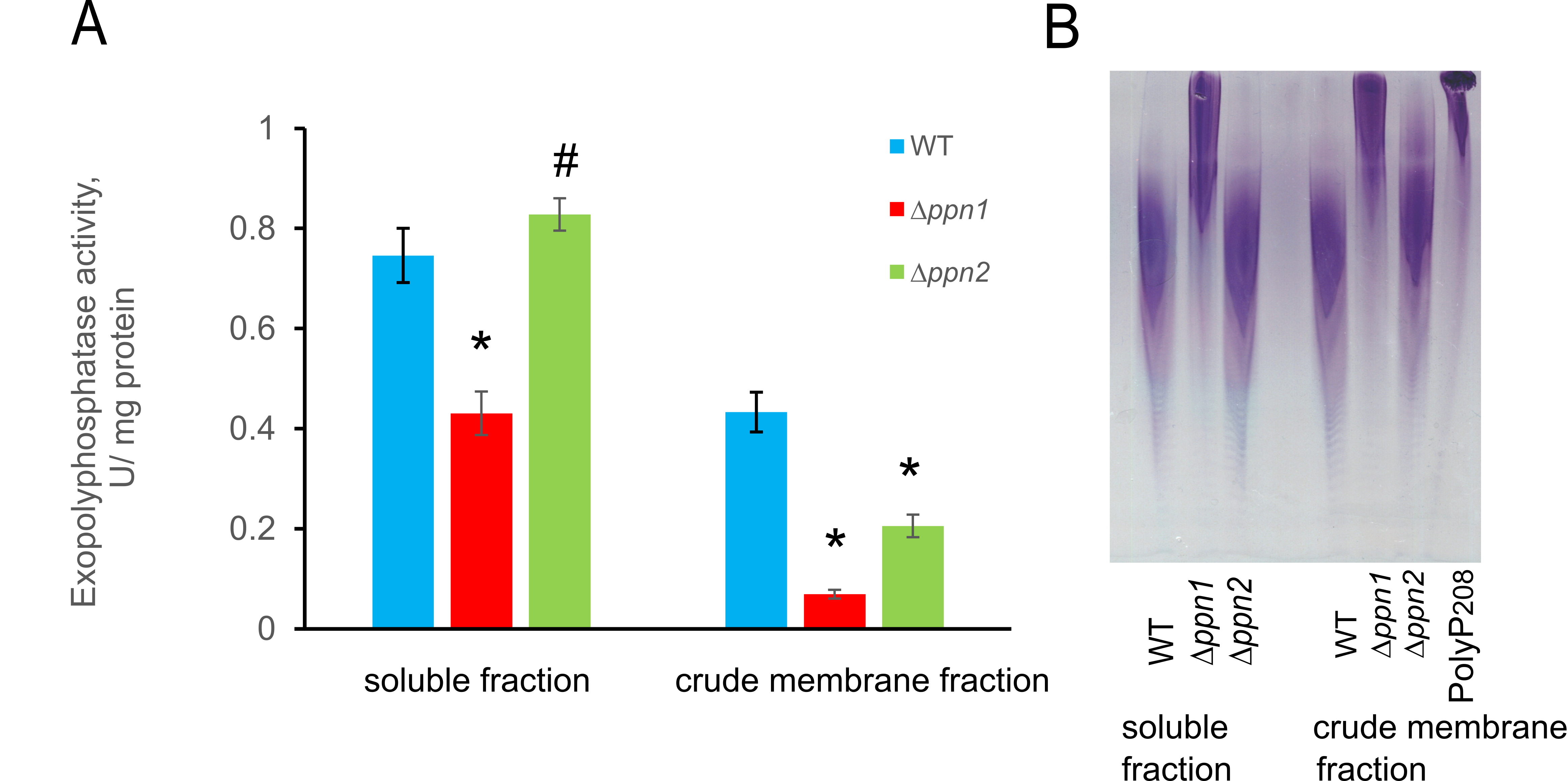
Figure 3. A - Exopolyphosphatase activities in the soluble and crude membrane fractions of polyP208. Statistical significance was assessed relative to the data for the parental wt strain with the help of Student's t-test, using the standard Excel program: * p < 0.001, # - the difference is statistically insignificant. B - Endopolyphosphatase activity (electropherograms of polyP208 after incubation with preparations of soluble and crude membrane fractions. PolyP208 - control incubation of polyphosphates with a chain length of about 208 phosphate residues without adding the studied samples.
These data are in agreement with the previous information about preferential localization of Ppn1 in the vacuolar and mitochondrial membranes and in the nuclei [13, 16]. Unexpectedly, the fraction of cell free extract obtained from the cells of the ∆ppn1 strain also demonstrated significantly reduced endo- and exopolyphosphatase activities (Fig. 3A and B). This fact suggests a considerable level of Ppn1 in the cytoplasm of the wt strain. This seems to be a characteristic feature of the BY4741 strain. Usually, the level of Ppn1 in the cytoplasm is low [15]. The method of obtaining cell free extract [16] used in the present work does not lead to the admixtures of cellular organelles. Previously, a significant increase in Ppn1 level in the cytoplasm was observed in the mutant lacking the Ppx1 polyphosphatase [11] and in the wild S. cerevisiae strain under the conditions of polyP super-accumulation ([15] or under DNA replication stress [21]. As regards the ∆ppn2 mutant, both exo- and endopolysphosphatase activities did not change in the cell free extract but decreased in the membrane fraction (Fig. 3). This is in agreement with the data on Ppn2 localization in the vacuolar membrane [13]. Ppn2 does not exhibit the exopolyphosphatase activity in vitro [14]. Apparently, the reduced ability to release Pi from polyP by the preparation of the crude membrane fraction of ∆ppn2 mutant is due to the fact that polyP fragmentation by endopolyphosphatase Ppn2 in the same fraction from the wt strain facilitates the subsequent hydrolysis by other enzymes such as pyrophosphatases. The decrease in polyphosphatase activity in both subcellular fractions is more pronounced for the ∆ppn1 mutant compared to the ∆ppn2 mutant. This is inconsistent with the fact that the increase in the content of long-chain polyP is more marked in the cells of the ∆ppn2 strain. Note, that the conditions in a cell may considerably different from those in vivo. Thus, the pH value inside the vacuoles is in the acidic range, whereas the optimum pH value for both polyphosphatases is in the neutral range. In addition, yeast cells may have other, currently unknown pathways for regulating polyP hydrolysis by polyphosphatases often localized together with polyP.
3.4. Polyphosphatase activity and polyP content in mitochondria
The inability of the ∆ppn1 mutant CRN [11] to grow on non-fermentable substrates such as lactate and ethanol was observed many years ago [17]. Its mitochondria were defective in respiration functions, lacked an exopolyphosphatase activity and possessed the doubled level of polyP in mitochondria [17]. The ∆ppn1 mutant used in this study had no growth defects when cultivated in the medium with ethanol (Fig. 1). To understand the differences between these two ∆ppn1 mutants of different origin, we obtained mitochondrial preparations from the wt and ∆ppn1 strains. The ATPase activities of the preparations were highly sensitive to a specific mitochondrial ATPase inhibitor, NaN3 (Table 1), indicating the sufficient purity of mitochondrial preparations from the admixtures of vacuoles and cytoplasmic membranes. Like in case of the previously studied pair of strains [17], the polyphosphatase activity in the mitochondrial preparation of the ∆ppn1 strain was insignificant (Table 1). However, the polyP content in mutant mitochondria only weakly increased during the growth on glucose (Table 2). As regards the preparation of mitochondria from the cells grown on ethanol, both the wt and ∆ppn1 strains showed a significant decrease in polyP content (Table 2), just as it was observed earlier in wild-type cells [22]. The polyP levels in ∆ppn1 mutant cells decreased during cultivation with ethanol despite the low level of the polyphosphatase activity. Apparently, there are other ways to regulate the polyP level in mitochondria, which are not implemented in the previously used CRN strain.
Table 1. ATPase and polyphosphatase activities in mitochondria preparations of S. cerevisiae strains WT and ∆ppn1.
|
Yeast strain |
ATPase, U/mg protein |
ATPase, inhibition with 5 mM NaN3, % |
Polyphosphatase, U/ mg protein |
|||
|
glucose |
ethanol |
glucose |
ethanol |
glucose |
ethanol |
|
|
WT |
0.53 ±0.01 |
0.51±0.048 |
80 |
87 |
0.58 ±0.02 |
0.055 ±0.001 |
|
∆ppn1 |
0.38 ±0.004 |
0.44 ±0.025 |
81 |
85 |
< 0.01 |
0.019 ±0.001 |
Table 2. Pi and polyP content (µmol P/mg protein) in mitochondria preparations of S. cerevisiae strains WT and ∆ppn1.
|
Yeast strain |
Pi |
Acid-soluble polyP
|
Acid-insoluble polyP |
|||
|
glucose |
ethanol |
glucose |
ethanol |
glucose |
ethanol |
|
|
WT |
0.14 ±0.02 |
0.069 ±0.001 |
0.31 ±0.06 |
0.12 ±0.01 |
0.52 ±0.003 |
0.34 ±0.004 |
|
∆ppn1 |
0.17 ±0.01 |
0.037 ±0.001 |
0.38 ±0.06 |
0.11 ±0.008 |
0.68 ±0.01 |
0.37 ±0.007 |
The results demonstrate that our previous hypothesis on the role of Ppn1 in the regulation of polyP level and formation of mitochondria from promitochondria [21] is incorrect, and the defects in mitochondrial function observed in the mutant CRN strain were caused by some unknown effects not connected with Ppn1.
3.5. Stress tolerance
Since polyP is involved in the mechanisms of stress resistance to heavy metal ions [4] and peroxide [23], the strains were compared with respect to their resistance to Cd2+, Mn2+ and peroxide.
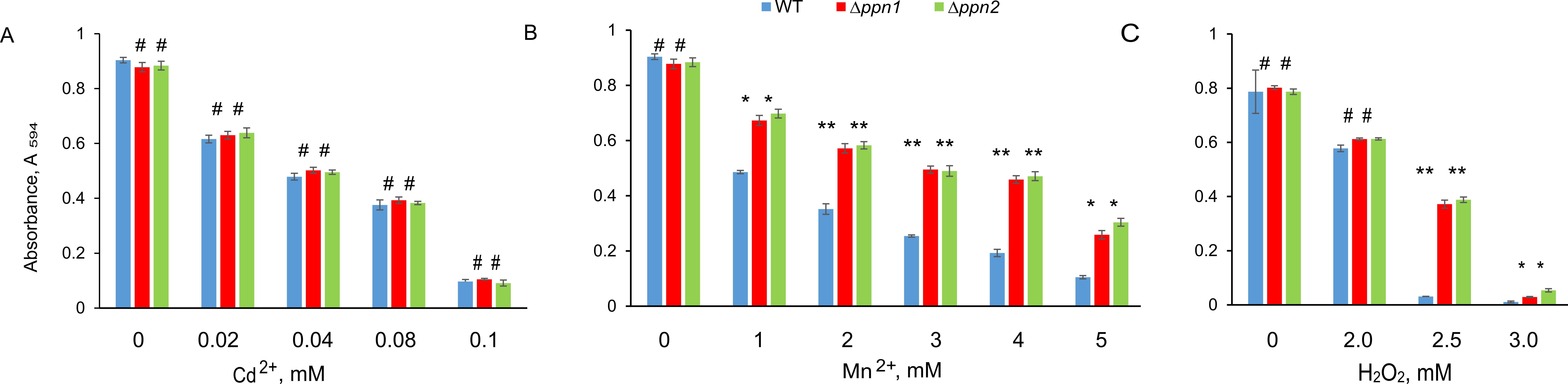
Figure 4. Effect of Cd2+, Mn2+ and hydrogen peroxide on the growth of wt, Δppn1 and Δppn2 strains of S. cerevisiae. The experiments were carried out in quadruplicate. Statistical significance was assessed relative to the data for the parental wt strain with the help of Student t-test using the standard Excel program: ** p < 10-4, * p < 0.001, # - the difference is statistically insignificant.
The resistance to Cd2+ was almost the same for all tested strains (Fig. 4A). Both ∆ppn1 and ∆ppn2 strains demonstrated a similar increase in resistance to Mn2+ (Fig. 4B) and hydrogen peroxide (Fig 4C). We believe that these changes are associated with the higher content of long-chained polyP.
Discussion
In this work we have shown that the knockout mutations in PPN1 and PPN2 genes lead to a combination of physiological effects observed at the stationary growth stage on glucose:
- Decreased polyphosphatase activity in the subcellular fraction containing membrane structures and organelles, including the mitochondrial fraction;
- Increased content of high molecular weight polyP, which is more marked in the PPN2 gene knockout mutant;
- Increased resistance to manganese and peroxide stress.
It can be assumed that polyP level under normal cultivation conditions are primarily determined by their synthesis, which is performed by the VTC complex [24]. Polyphosphatases with the endopolyphosphatase activity such as Ppn1 and Ppn2 are involved in the reduction of polyP chain length, which is probably needed for distribution of the resultant polyP across organelles and compartments such as mitochondria, nuclei and cytoplasm. Although many authors considered vacuoles to be the main polyP-containing compartments in yeast [13], it was shown that polyPs were present in the nuclei, cytoplasm, and mitochondria [16]. PolyP elongation in the ∆ppn1 and ∆ppn2 mutants is likely to influence various cellular processes, including the regulation of enzyme activity, as well as the ability to bind toxic metal ions. Data on the effect of these mutations on the total amount of polyP in the cell are ambiguous. Thus, one publication reported that the polyP content increased in the cells of the ∆ppn1 mutant, but not in ∆ppn2 [26], while other authors [7] showed that even in the ∆ppn1ppn2ppx1ddp1 mutant, the total polyP content differed little from that in wild-type cells [7]. In our case, the total polyP content in the mutants did not differ from that in the parent strain. Of primary interest is the increase in the content of long-chain polyP, which was noted both in the present work and in the works of other authors [7, 26]. At the moment, it is not possible to trace a direct relationship between this change and the numerous pleiotropic effects of the ∆ppn1 and ∆ppn2 mutations. Recently, new effects of these mutations have been identified: both mutants have a reduced replicative lifespan [26], cells of the Δppn1Δppx1Candida albicans mutant are characterized by increased sensitivity to stress and pseudohyphal morphology [27]. It can be assumed that the multiple effects of these mutations may be associated with both the regulatory role of polyP, able to directly interact with certain regions of DNA [9], and with the ability of Ppn1 and Ppn2 to participate in the cleavage of polyP from lysine-polyphosphorylated proteins [7] or to hydrolyze various signaling molecules [14].
The results of this work show that one of the previously observed effects of the Δppn1 mutation, namely, the defect in mitochondrial development and the inability to grow on non-fermentable carbon sources [17] in the CRN mutant for the PPN1 gene constructed by Sethuraman et al. [11], is not reproduced in the commercially available Δppn1 strain we used. The data on the determination of polyphosphate activity indicate that this strain indeed lacks the Ppn1 polyphosphatase. Therefore, our previous hypothesis about the role of Ppn1 in the regulation of polyP levels and the formation of mitochondria from promitochondria [17] is incorrect, and the defects in mitochondrial function observed in the CRN mutant strain are not associated with the absence of Ppn1, but are most likely due to damage to other genes during strain construction. Note that the genotype of the parent strain BY4741 (MATahis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) differs from the genotype of the parent strain CRY (MATaade2 his3 leu2 trp1 ura3), which was used by the authors [11] to construct the mutant CRN ∆ppn1 (MATa ade2 his3 ura3 ppn1∆::CgTRP1).
The mutants lacking polyphosphatases Ppn1or Ppn2 showed an increase in polyP chain length and also the enhanced resistance to manganese and peroxide. The Ppn1-overexpressing strain also showed a higher resistance to both types of stresses, while the Ppn2-overexpressing strain was more resistant to peroxide [25]. Apparently, the pathways leading to this increase are different in case of knockout mutation and overexpression of these genes. In knockout mutations, the increase in manganese resistance can be explained by the elevated level of high-molecular-weight polyPs and a better binding of these ions, while in case of overexpression, when the level of high-molecular-weight polyPs decreases, more complex indirect pathways can be assumed including also those associated with the influence on the level of such secondary signaling molecules as inositol polyphosphates, whose metabolism is closely linked to the metabolism of polyP [12].
Author contributions
Trilisenko L.V., Ledova L.A., Ryazanova L.P., Kulakovskaya E.V., Tomashevsky A.A. – implementation and analysis of experiments, Kulakovskaya T.V. – writing the manuscript. All authors have read and agreed to the published version of the manuscript.
Conflict of interest
The authors declare no conflict of interest.
References
- Rao N. N., Gómez-García M. R. and Kornberg A. Inorganic polyphosphate: Essential for growth and survival. Ann Rev Biochem., 78, 605–647 (2009).
- Kus F., Smolenski R. T. and Tomczyk M. Inorganic polyphosphate-regulator of cellular metabolism in homeostasis and disease. Biomedicines, 10 (4), 913 (2022). DOI: 10.3390/biomedicines10040913.
- McCarthy L. and Downey M. The emerging landscape of eukaryotic polyphosphatases. FEBS Lett., 597 (11), 1447–1461 (2023). DOI: 10.1002/1873-3468.14584.
- Kulaev I. S., Vagabov V. M. and Kulakovskaya T. V. The biochemistry of inorganic polyphosphates, (New York USA, Wiley, 2004).
- Bru S., Martínez-Laínez J. M., Hernández-Ortega S., Quandt E., Torres-Torronteras J., Martí R., Canadell D., Ariño J., Sharma S., Jiménez J. and Clotet J. Polyphosphate is involved in cell cycle progression and genomic stability in Saccharomyces cerevisiae. Mol Microbiol., 101 (3), 367–380 (2016). DOI: 10.1111/mmi.13396.
- Kalebina T. S., Egorov S. N., Arbatskii N. P., Bezsonov E. E., Gorkovskii A..A. and Kulaev I. S. The role of high-molecular-weight polyphosphates in activation of glucan transferase Bgl2p from Saccharomyces cerevisiae cell wall. Dokl Biochem Biophys., 420, 142–145 (2008). DOI: 10.1134/s1607672908030125.
- Azevedo C., Desfougères Y., Jiramongkol Y., Partington H., Trakansuebkul S., Singh J., Steck N., Jessen H. J. and Saiardi A. Development of a yeast model to study the contribution of vacuolar polyphosphate metabolism to lysine polyphosphorylation. J Biol Chem,. 295 (6), 1439–1451 (2020). DOI: 10.1074/jbc.RA119.011680.
- McCarthy L., Bentley-DeSousa A., Denoncourt A., Tseng Y .C., Gabriel M. and Downey M. Proteins required for vacuolar function are targets of lysine polyphosphorylation in yeast. FEBS Lett., 594 (1), 21–30 (2020). DOI: 10.1002/1873-3468.13588.
- Sanchez A. M., Garg A., Schwer B. and Shuman S. Inorganic polyphosphate abets silencing of a sub-telomeric gene cluster in fission yeast. Micro Publ Biol., 3; (2023) DOI: 10.17912/micropub.biology.000744. DOI: 10.17912/micropub.biology.000744.
- Wurst H., Shiba T. and Kornberg A. The gene for a major exopolyphosphatase of Saccharomyces cerevisiae. J Bacteriol., 177: 898–906 (1995). DOI: 1128/jb.177.4.898-906.1995.
- Sethuraman A., Rao N. N., and Kornberg A. The endopolyphosphatase gene: essential in Saccharomyces cerevisiae, Proc Natl Acad Sci USA, 98 (15), 8542–8547 (2001). DOI: 10.1073/pnas.151269398.
- Lonetti A., Szijgyarto Z., Bosch D., Loss O., Azevedo C. and Saiardi A. Identification of an evolutionarily conserved family of inorganic polyphosphate endopolyphosphatases, J Biol Chem., 286, 31966–31974 (2011). DOI: 10.1074/jbc.M111.266320.
- Gerasimaitė R. and Mayer A. Ppn2, a novel Zn2+-dependent polyphosphatase in the acidocalcisome-like yeast vacuole. J Cell Sci., 130 (9), 1625–1636 (2017). DOI:1242/jcs.201061.
- Andreeva N., Ledova L., Ryazanova L., Tomashevsky A., Kulakovskaya T. and Eldarov M. Ppn2 endopolyphosphatase overexpressed in Saccharomyces cerevisiae: comparison with Ppn1, Ppx1, and Ddp1 polyphosphatases. Biochimie, 163, 101–107 (2019). DOI: 10.1016/j.biochi.2019.06.001.
- Andreeva N. A., Kulakovskaya T. V., and Kulaev I. S. High molecular mass exopolyphosphatase from the cytosol of the yeast Saccharomyces cerevisiae is encoded by the PPN1 Biochemistry (Moscow), 71 (9),975–977 (2006). DOI: 10.1134/s0006297906090045.
- Lichko L., Kulakovskaya T., Pestov N. and Kulaev I. Inorganic polyphosphates and exopolyphosphatases in cell compartments of the yeast Saccharomyces cerevisiae under inactivation of PPX1 and PPN1 Biosci Rep., 26 (1), 45–54 (2006). DOI: 10.1007/s10540-006-9003-2.
- Pestov N. A., Kulakovskaya T. V. and Kulaev I. S. Effects of inactivation of the PPN1 gene on exopolyphosphatases, inorganic polyphosphates and function of mitochondria in the yeast Saccharomyces cerevisiae. FEMS Yeast Res., 5 (9), 823–828 (2005). DOI: 10.1016/j.femsyr.2005.03.002.
- Vagabov V. M., Trilisenko L. V. and Kulaev I. S. Dependence of inorganic polyphosphate chain length on the orthophosphate content in the culture medium of the yeast Saccharomyces cerevisiae. Biochemistry (Moscow), 65, 349–355 (2000).
- Heinonen J. K. and Lahti R. J. A new and convenient colorimetric determination of inorganic orthophosphate and its application to the assay of inorganic pyrophosphatase. Anal Biochem., 113 (2), 313–317 (1981). DOI: 10.1016/0003-2697(81)90082-8.
- Zinzer E. and Daum G. Isolation and biochemical characterization of organelles from the yeast Saccharomyces cerevisiae. Yeast, 11, 493–536 (1995). DOI: 10.1002/yea.320110602.
- Tkach J. M., Yimit A, Lee A. Y., Riffle M., Costanzo M., Jaschob D., Hendry J. A., Ou J., Moffat J., Boone C., Davis T. N, Nislow C. and Brown G. W. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat Cell Biol., 14 (9), 966–976 (2012). DOI: 10.1038/ncb2549.
- Andreeva N. A., Kulakovskaya T. V., Kulakovskaya E. V. and Kulaev I. S. Polyphosphates and exopolyphosphatases in cytosol and mitochondria of Saccharomyces cerevisiae during growth on glucose or ethanol under phosphate surplus. Biochemistry (Moscow), 73 (1), 65–69 (2008). DOI: 10.1134/s0006297908010094.
- Culotta V. C. and Daly M. J. Manganese complexes: diverse metabolic routes to oxidative stress resistance in prokaryotes and yeast. Antioxid Redox Sign, l19 (9), 933–944 (2013). DOI: 10.1089/ars.2012.5093.
- Hothorn M., Neumann H., Lenherr E.D., Wehner M., Rybin V., Hassa P.O., Uttenweiler A., Reinhardt M., Schmidt A., Seiler J., Ladurner A. G., Herrmann C., Scheffzek K. and Mayer A. Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science, 324 (5926), 513–516 (2009). DOI: 10.1126/science.1168120.
- Andreeva N., Ryazanova L., Ledova L., Trilisenko L. and Kulakovskaya T. Stress resistance of Saccharomyces cerevisiae strains overexpressing yeast polyphosphatases. Stresses, 2, 17–25 (2022). DOI: 10.3390/Stresses2010002.
- Umeda C., Nakajima T., Maruhashi T., Tanigawa M., Maeda T. and Mukai Y. Overexpression of polyphosphate polymerases and deletion of polyphosphate phosphatases shorten the replicative lifespan in yeast. FEBS Lett., 597 (18), 2316-2333 (2023). DOI: 10.1002/1873-3468.
- Ahmed Y., Ikeh M. A. C., MacCallum D. M., Day A. M., Waldron K. and Quinn J. Blocking polyphosphate mobilization inhibits pho4 activation and virulence in the pathogen Candida albicans. mBio., 13 (3), e0034222 (2022). DOI: 10.1128/mbio.00342-22.
License
Copyright (c) 2024 Трилисенко Л. В. , Ледова Л. А. , Рязанова Л. П. , Екатерина Кулаковская, Томашевский А. А. , Кулаковская Т. В. (Автор)

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.